Weekly Links #9
Automating gene-editing experiments. Computational design of protein nanomaterials. The end of scientific empires.
A healthy baby was born from an embryo that was frozen in 1994.
Automation of gene-editing experiments with CRISPR-GPT.
Computational design of protein nanomaterials.
A GFP-expressing mouse model to test the efficiency of in vivo gene editing tools. Looks genuinely useful.
An autonomous device that generates “an active protein from two otherwise inactive polypeptide fragments” using trans-splicing.
Review on AI for gene-editing. Add crypto, profit.
An AI-generated, open-source version of Cas9. This paper was published on bioRxiv more than a year ago, but it’s finally out in Nature. Congrats to the Profluent team!
A “multichannel bioelectronic sensor where different chemicals regulate distinct extracellular electron transfer pathways within a single Escherichia coli cell.” Basically, using a single cell as a whole-cell biosensor for two different molecules. Neat idea.
DynaTag is a tool to map transcription factors, and the DNA sequences to which they band, at single-cell resolution and in small volumes!
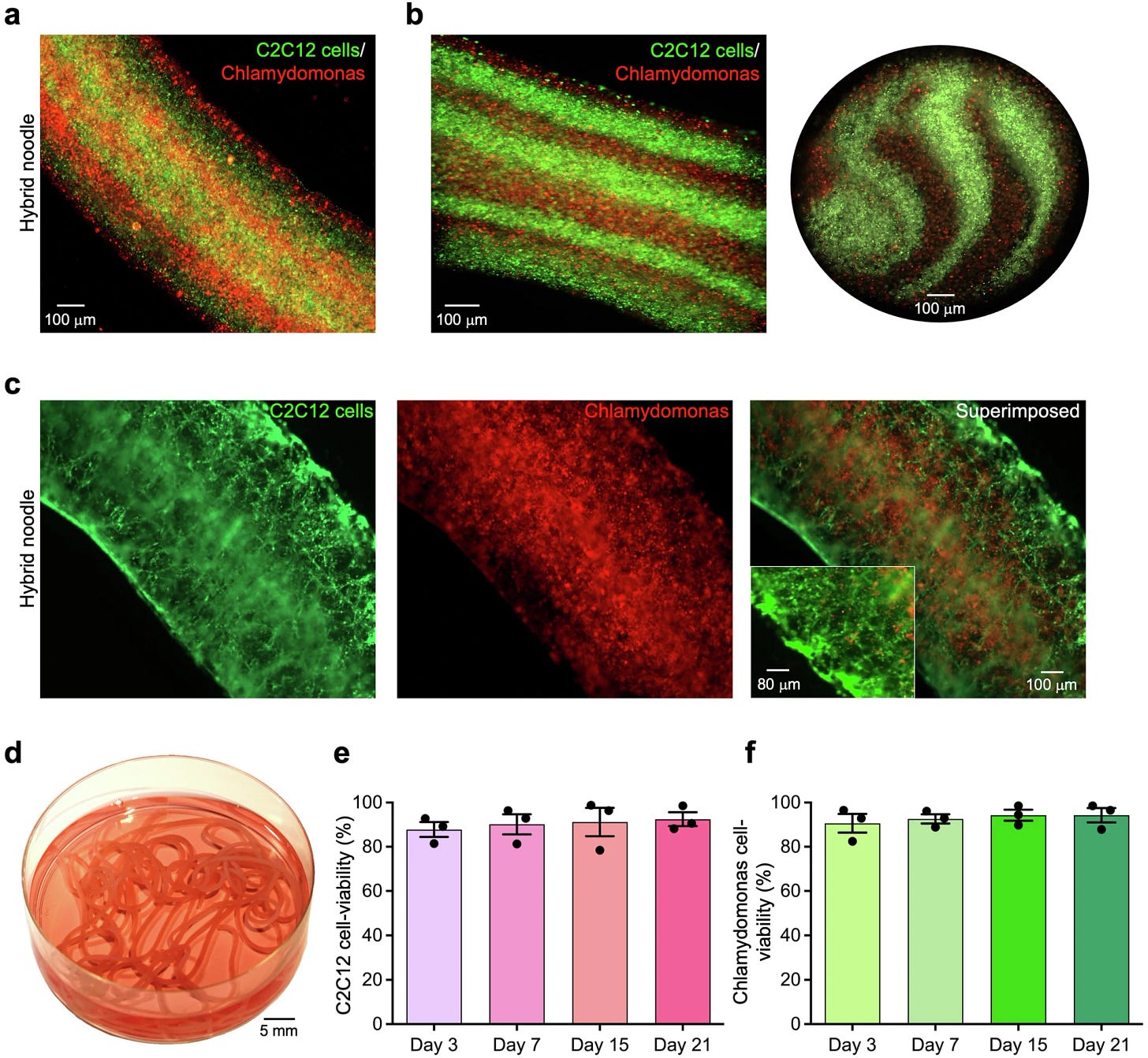
These noodles were made by 3D printing a mixture of plant and animal cells. I’ll try it if you try it.
Can an individual cell remember past experiences? Nice article from Quanta.
A minimal chemotactic cell. Researchers took a vesicle, filled it with a single type of enzyme and some protein pores, and showed that this "minimal cell," made from just three components (!!), could "actively propel itself toward an enzyme substrate gradient." See my writeup on X.
A “virtual lab of AI agents” design nanobodies against SARS-CoV-2. My general take on this kind of thing is that it’s not super important, and we’re going to see a lot of it for any domain in which the general steps toward success are well-known. So making nanobodies, running gene-editing experiments, analyzing a particular type of dataset, searching papers for statistical anomalies, and so on….all can be automated.
Synthetic biology + ingestible electronics. “…we engineered E. coli Nissle 1917 to detect inflammation-associated nitric oxide in the pig gut and generate a bioluminescent signal for diagnosis of colitis. This signal is transduced by the optoelectronic capsule into a wireless electrical signal and remotely monitored by a smartphone.”
Engineering a plant immune receptor to defend crops against a wide range of pathogens. See my writeup on X.
Ångström-resolution imaging of cell-surface glycans.
Engineering proteins to “form micro-to millimeter scale structures” in cell-free liquids.
“…we introduce a framework that integrates orthogonal operational amplifiers (OAs) into standardized biological processes to enable efficient signal decomposition and amplification. By engineering σ/anti-σ pairs, varying ribosome binding site (RBS) strengths, and utilizing both open-loop and closed-loop configurations, we design scalable OAs that enhance the precision, adaptability, and signal-to-noise ratio of genetic circuits.”
“The modern-day potato evolved from the hybridization of the ancestors of tomato plants, and another potato-like plant, in South America.” So basically potatoes are part tomato. Original paper in Cell.
How do yeast figure out which “pathway” to use to secrete their proteins? The length of a hydrophobic helix in the signal peptide seems to matter quite a bit.
Two mRNA-based vaccine papers, phase I and preclinical, for HIV. #1 and #2.
Metascience is more important now than ever
Every scientific empire comes to an end. We need more odes to American dynamism!
What is the optimal amount of time to give antibiotics to people with a blood infection? It doesn’t seem to make much difference.
An “effective chemical method” to boost the rate at which cells release outer membrane vesicles by up to 65-fold.
Computational design of “binder” proteins that can grab onto intrinsically disordered proteins.
Using nanopores to sequence DNA with non-canonical bases! “We demonstrate that [noncanonical bases] can be robustly sequenced on a MinION system ( > 2.3×106 reads/flowcell)…”
Ants as liquid brains. “…a simple feedback mechanism, mediated by local interactions, governs the foraging patterns of A. senilis. Such feedback is modulated by adjusting the proportion of two coexisting movement behaviors: recruits, which facilitated information transfer and food exploitation by aggregating closely to the nest and the food patches, and scouts, which could bypass this feedback and discover alternative food sources. Therefore, distinct movement patterns contributed differently to optimizing each phase of the foraging process, proving an adaptive mechanism to balance exploration and exploitation.”
A “proof of concept for atomic precision vaccination” against HIV in humans, using a stabilized version of the Env trimer to induce B cell responses.
An Escherichia coli with a 57-codon genetic code. The synthetic organism is called Syn57 and it uses just 55 codons to encode the 20 canonical amino acids, along with a start and stop codon. The full synthetic genome is 4 million bases long, a roughly 20 percent reduction from natural E. coli genomes.
The deepest, chemosynthetic community of lifeforms ever discovered. This paper reports elongated worms (called "siboglinids") thriving at depths of 9,500 meters. At that depth, the pressure exerted on these organisms is about 943 atmospheres or 95.6 MPa.
Using directed evolution to turn cytochrome P450 enzymes into enzymes that can perform a new reaction not found in nature: metal-hydride hydrogen atom transfer.
Researchers made a tiny, molecular machine (powered by light) that twists strings of atoms.
Sponsored by Asimov Press. Subscribe!